Background
This path describes the process of authoring a pathway model from a published pathway figure from the Pathway Figure OCR resource. Pathway Figure OCR (PFOCR) is an open science project dedicated to extracting pathway information from the published literature. These instructions assume that you have everything you need to work with WikiPathways, including PathVisio and a WikiPathways account.
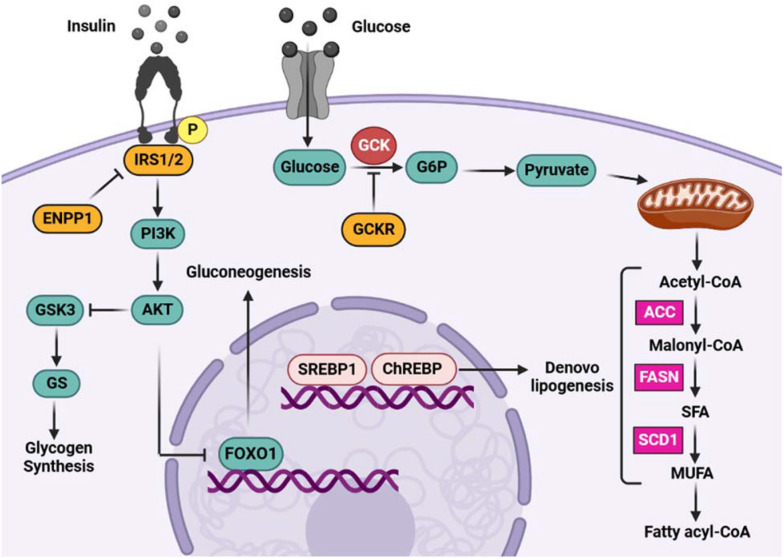
We will focus on the pathway figure below, describing Glucose-Insulin signaling, from a publication by Mahmoudi et al. The paper describes the genetic variation on non-alcoholic fatty liver disease and the connection to the Glucose-Insulin signaling pathway. To model this pathway, we will use the corresponding Pathway Figure OCR page as well as the publication, and other resources. The first step, described below, is to access the gene products and metabolites extracted by PFOCR to jump-start the pathway drawing process.
Getting Started
- Launch PathVisio and set up identifier mapping for human.
- Start a new pathway by selecting File > New. Close the Pathway properties interface, and then reopen it by double-clicking on the Pathway Information area at the top left.
- Enter a pathway title, for example "Glucose-insulin Signaling Pathway", and select "Homo sapiens" as Organism.
- Navigate to the Pathway Figure OCR webpage.
- Scroll down to Gene mentions and click the curved arrow symbol
 and select Copy DataNodes for PathVisio.
and select Copy DataNodes for PathVisio. - In PathVisio, select Edit > Paste to paste the nodes. This will produce a set of stacked data nodes, that are already annotated with external references.
- Repeat the process for the Chemical mentions on the PFOCR page. The metabolite nodes may end up in the same location as the gene product nodes; if so move the stack of nodes by click-and-drag while they are still selected.
- The information from PFOCR is not always accurate, meaning words in the figure may be matched incorrectly to gene and chemical annotations. Because of this, the data node annotation needs to be double-checked, and corrections made. In our case, some of the data nodes are false positives and should be deleted in PathVisio. Go ahead and delete the following nodes: MAP4K2, FAM20C, MAP4K5, GNAS, GNAL, BMS1, ACC (metabolite) and Glycogen.